We’re open-sourcing mBER, our breakthrough AI library for designing epitope-specific antibodies. To showcase its power, we conducted the largest reported de novo antibody experiment—testing more than 1 million designs against 145 diverse targets in an experiment measuring over 100 million protein–protein interactions. The study identified specific binders to around half of targets tested, with success rates up to 40% on optimal epitopes, on par with state-of-the-art approaches. We built mBER to leverage the massive scale of our in vivo multiplexing technology for screening millions of designs directly in living systems. Together, these form the foundation of Manifold’s direct-to-vivo drug discovery platform, bridging the gap between AI-guided design and biological translation. We are already deploying mBER to advance a pipeline of tissue-targeted medicines, while using the data to build organism-scale models of biology.
Overview
Artificial intelligence is increasingly transforming drug discovery, dramatically improving on trial-and-error methods. But most of the iteration is informed by testing against cells in a dish (in vitro). The ultimate measure of success is improving outcomes in patients, where failure rates remain unacceptably high. Unlocking the full potential of AI requires solving a translation problem: drugs must work in people but they are designed in reductionist systems.
The vision for Manifold Bio is to realize an approach to drug discovery that drastically increases experimental throughput and AI-guided iteration in living (in vivo) systems. This brings us closer to human biology, and a big step toward the long‑term goal of zero‑shot drug design: going from a desired therapeutic profile to a molecule that just works in patients.
Machine Learning (ML) alone will not achieve this vision. The key is ML2: “Machine Learning with Multiplexed Libraries”–Manifold’s approach to engineering biological systems that combines ML-guided design with massively parallel molecular synthesis and measurement technologies.
Leveraging ML2, Manifold has built the first direct-to-vivo drug discovery platform, positioning us to unlock the full transformational potential of AI-guided protein design. With the unique technological moat to test millions of binder designs directly in living systems, we are able to rapidly generate organism‑scale evidence on biodistribution, cell‑type engagement, and functional outcomes—providing the biologically relevant, high-quality data needed for model training and predictive design.
mBER was developed to harness the scale of our in vivo screening capabilities: a de novo binder generator capable of producing millions of designs across thousands of targets. mBER plays a crucial role in Manifold’s mission, generating quality design hypotheses at scale, while multiplexed in vivo readouts provide high-value labels. Together, these capabilities form an unprecedented flywheel for learning from data and building generalizable maps from drug design to therapeutic effect.
What is mBER?
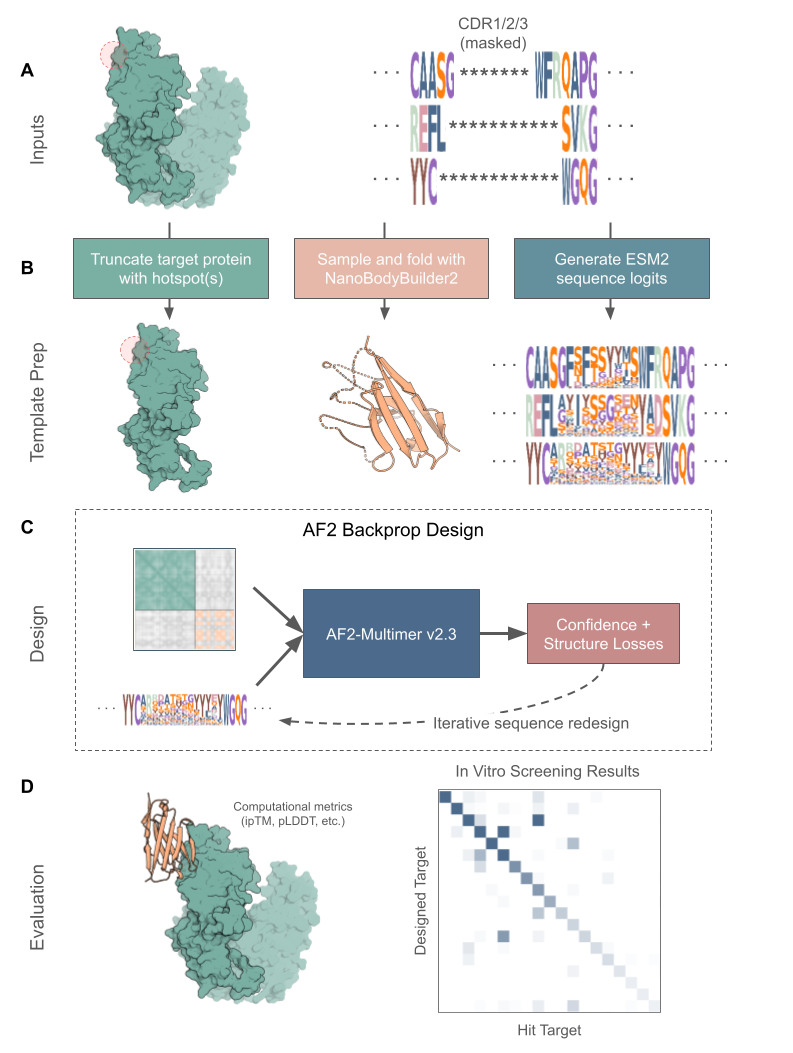
mBER is a protein design framework that enables antibody binder design by conditioning AlphaFold-Multimer with sequence and structure information for any desired antibody framework. mBER directly designs and optimizes antibody sequences by inverting the AlphaFold network—iteratively updating an input sequence until AlphaFold is confident in its bound structure against a target epitope of interest.
While tools like ColabDesign and BindCraft pioneered and demonstrated the potential of backpropagation, they produce unconstrained “minibinders” that lack the developability advantages of antibodies. mBER adds two key innovations:
- Structural templates that ensure designs respect antibody architecture
- Sequence priors from protein language models that bias variable regions toward natural human antibody sequences.
This combination enables generation of realistic and experimentally successful antibody binders without any retraining of underlying folding models! We find that AlphaFold’s confidence metrics, particularly ipTM, are well aligned with experimental success. By optimizing sequences with respect to AlphaFold’s confidence metrics and filtering out designs with low-confidence folds, we are able to achieve state-of-the-art experimental success rates for de novo antibody binder design with mBER.
100M-scale all-against-all experiment validates mBER
To validate mBER and exemplify the power of ML2, we designed over one million VHH antibodies against 436 human receptors and validated them in a pair of experiments where we tested 1,153,241 binders against 145 targets. By measuring a designed antibody against its intended target and every other target we can confidently validate its binding and specificity. In total these experiments measured over 100 million antibody-target interactions. We found specific binders to nearly half our targets (65 of 145), with success rates reaching 40% on optimal epitopes after computational filtering. This represents the largest reported de novo antibody validation campaign.
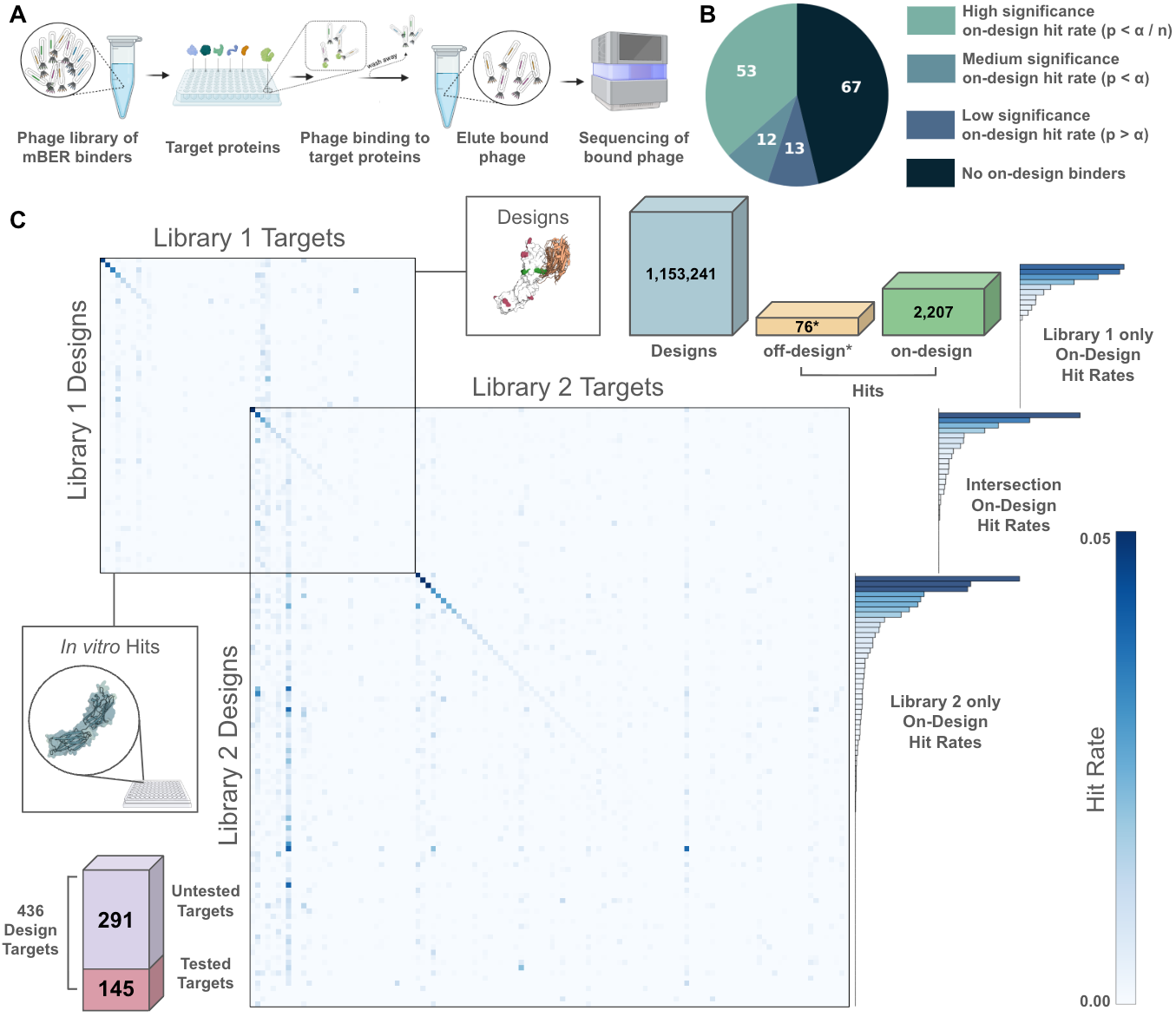
We report the details of this design and experimental validation in a preprint here: biorxiv.org/content/10.1101/2025.09.26.678877v1
Why generate millions of antibodies to hundreds of targets?
We built direct-to-vivo and mBER to solve real therapeutic challenges that demand scale.
One of our first focus areas is creating medicines that can cross the blood-brain barrier (BBB)--a grand challenge where most progress has been around exploiting a single receptor, Transferrin Receptor (TfR), whose promise and limitations are becoming better understood. However, there are hundreds of “portals”, receptors that could facilitate entry into the brain, and Manifold is uniquely positioned to systematically search the space of molecular shuttles against the full space of putative portals. mBER makes this possible by generating vast libraries of precise designs. With our multiplexed in vivo platform, we can screen all of these candidates at once, directly in living systems.
Beyond the brain, hundreds of medicines are held back by the challenge of tissue-specific delivery beyond the liver. We have expanded our efforts toward the ambitious mission of solving delivery to all tissues and cell types. To this end, we are building the EpiTome—a proprietary data resource linking epitopes to in vivo biodistribution and a toolbox of binders ready to advance into medicines. We’re extending mBER to systematically design binders covering epitopes of every known cell surface receptor, and advancing these into in vivo screens.
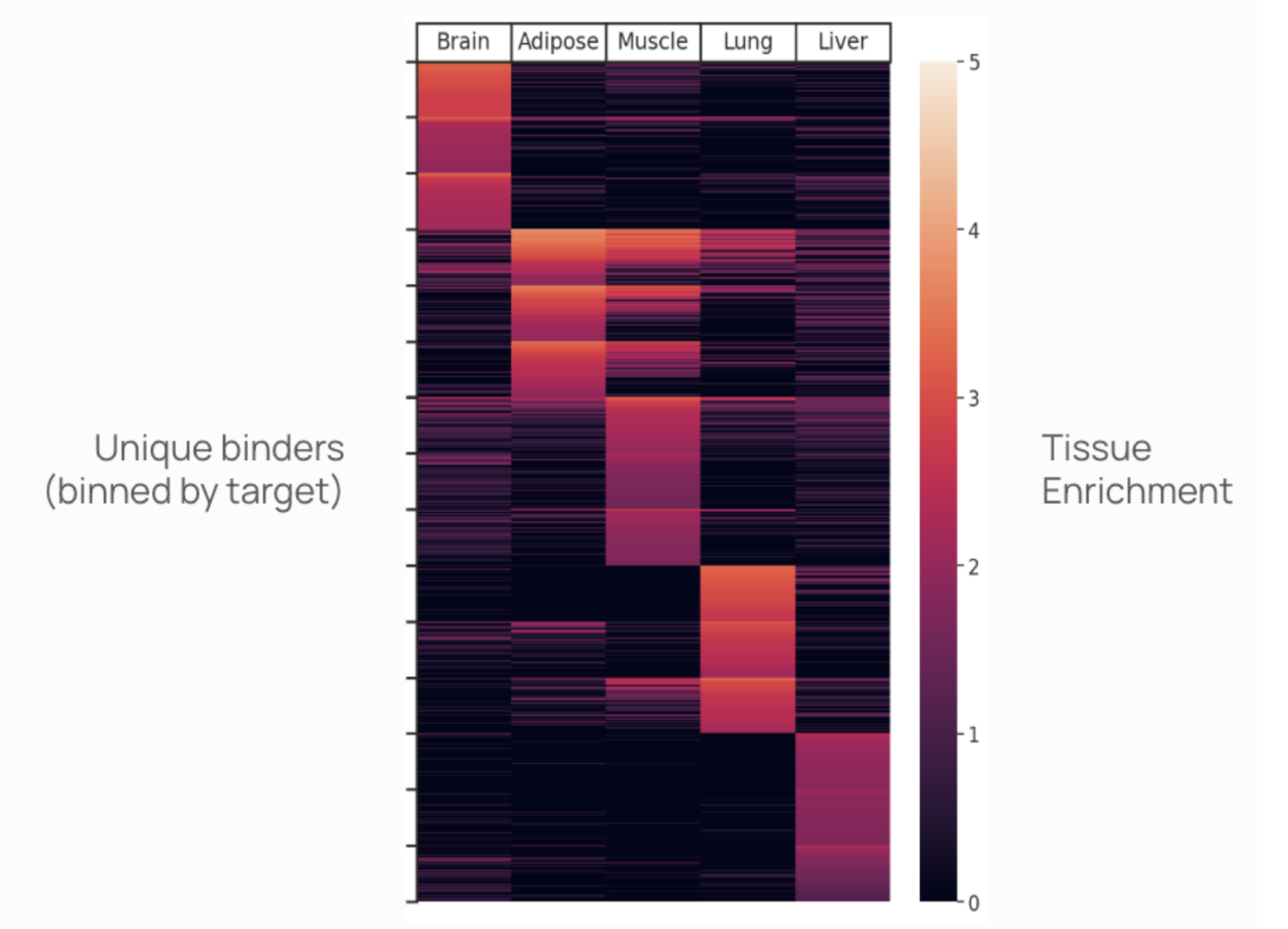
A final benefit of our comprehensive design and screening efforts is an unprecedented opportunity to expand the corpus of protein structure data. Public protein structure databases contain just a couple hundred thousand structures painstakingly produced by the collective efforts of the scientific community over decades. At Manifold, by generating millions of synthetic structures and then running massively-parallel experimental validation of each one, we are building a rich collection of labeled, semi-synthetic structure data. This includes direct-to-vivo validation of binders to targets that are difficult, if not impossible, to express in vitro. We believe this type of data will make up the bulk of inputs to the training of the next-generation of protein folding models. Better folding models will produce better designs, which will in turn increase the data generation capabilities of experiments. This virtuous cycle will accelerate from here.
The future of drug discovery
Together, mBER and our multiplexed in vivo screening technologies are allowing us to reimagine therapeutic discovery:
- Design at scale: Millions of epitope-specific binders with structural priors.
- Validation at scale: All tested directly in vivo in just a few experiments.
- Learning at scale: Mapping the EpiTome and fueling foundation models of biology.
This is how we will solve the hardest problems in medicine, starting with brain delivery and expanding to tissue targeting across the body.
Join Us
We're open-sourcing mBER because progress accelerates when the community can build together. You can:
- Use it: Design antibodies for your targets (github.com/manifoldbio/mber-open)
- Extend it: Contribute better priors, losses, and evaluation methods
- Partner with us: Create targeted medicines together (bd@manifold.bio)
mBER and the EpiTome are the beginning. We're building toward programmable medicines—where you specify the tissue, cell type, and biological effect, and AI designs the molecule to achieve it. This future requires massive biological data, powerful models, and a new approach to drug discovery.
Oh, and we’re hiring: We’re expanding our AI and platform teams. We’re already one of the most data-rich places in the world to build the next generation of models and measurement technologies, and this is only accelerating.
We're excited to build with you.